Interview with Onur Boyar: Drug and material design using generative models and Bayesian optimization
In this interview series, we’re meeting some of the AAAI/SIGAI Doctoral Consortium participants to find out more about their research. Onur Boyar is a PhD student at Nagoya university, working on generative models and Bayesian methods for materials and drug design. We met Onur to find out more about his research projects, methodology, and collaborations […]


In this interview series, we’re meeting some of the AAAI/SIGAI Doctoral Consortium participants to find out more about their research. Onur Boyar is a PhD student at Nagoya university, working on generative models and Bayesian methods for materials and drug design. We met Onur to find out more about his research projects, methodology, and collaborations with chemists.
Could you start by giving us a quick introduction, where you’re studying and the broad topic of your research?
I’m from Turkey, and I came to Japan three years ago to pursue my PhD. Before coming here, I was already interested in generative models, Bayesian methods, and Markov chain Monte Carlo techniques. Since Japan has a strong reputation in bioinformatics and the intersection of AI and the life sciences, I was eager to explore applied research in bioinformatics-related problems. My professors suggested working on drug and material design using generative models and optimization techniques. It turned out to be a great fit for me, especially because this research area heavily relies on Bayesian methodologies, which I wanted to delve deeper into. Since 2022, I’ve been working on optimization and generative methods for drug discovery. Along the way, we’ve developed several novel methodologies for both molecular and crystal material design.
Could you tell us about the projects that you’ve been working on during your PhD?
So, one of the main projects I’ve been working on during my PhD is part of Japan’s big Moonshot program — it’s a government-funded initiative with a pretty ambitious goal. The idea is to eventually build an AI scientist robot that can handle the entire drug discovery and development process on its own. Our team is focusing on developing the AI part of that system. The first step is to complete the drug development cycle in collaboration with chemists.
That’s the main project, but I’m also involved in a few others. For instance, we recently started collaborating with some chemists on drug discovery for a specific disease. We’re also working with a couple of companies on materials science problems, using our methods for generating crystal materials.
Each project has its own challenges and requires a slightly different approach. Early on, when we generated candidate molecules for drug discovery, the chemists told us they were struggling to actually synthesize the molecules because AI tends to produce structures that are hard to interpret or work with in practice. So, they asked if we could develop a method that edits existing molecules instead of generating entirely new ones. That way, they’d be working with structures they’re already familiar with, but slightly optimized for better properties. That shift in approach really helped — and we’ve seen some success with it. A few of the molecules we designed have shown promising properties, and some are even going through animal testing now. Drug development is a long and complex process, but we’re definitely making progress.
Is there one particular project that you’ve particularly enjoyed working on?
Yeah, actually, it’s the project I just mentioned. It’s been the most exciting one for me because I get to see real-world impact. I send over the molecules we generate, and the chemists actually try to synthesize them — that part feels really rewarding. It makes me feel like I’m genuinely contributing to science. It’s also great to collaborate closely with chemists, getting their feedback, seeing which molecules they find promising, and knowing that some are actually being produced. That kind of hands-on, practical side of the work is what I enjoy the most.
Could you tell us a bit more about your methodology for this project?
Sure! So first, we had a lot of in-depth discussions with my PhD supervisor, advisors, and our chemist collaborators. They were very clear about which parts of the molecules they wanted to preserve and what kinds of outcomes would be most useful for them. Based on their input, my main goal was to develop a sample-efficient methodology. Generating and testing thousands of molecules might be possible in theory, but in practice, it’s just not feasible due to time and budget constraints.
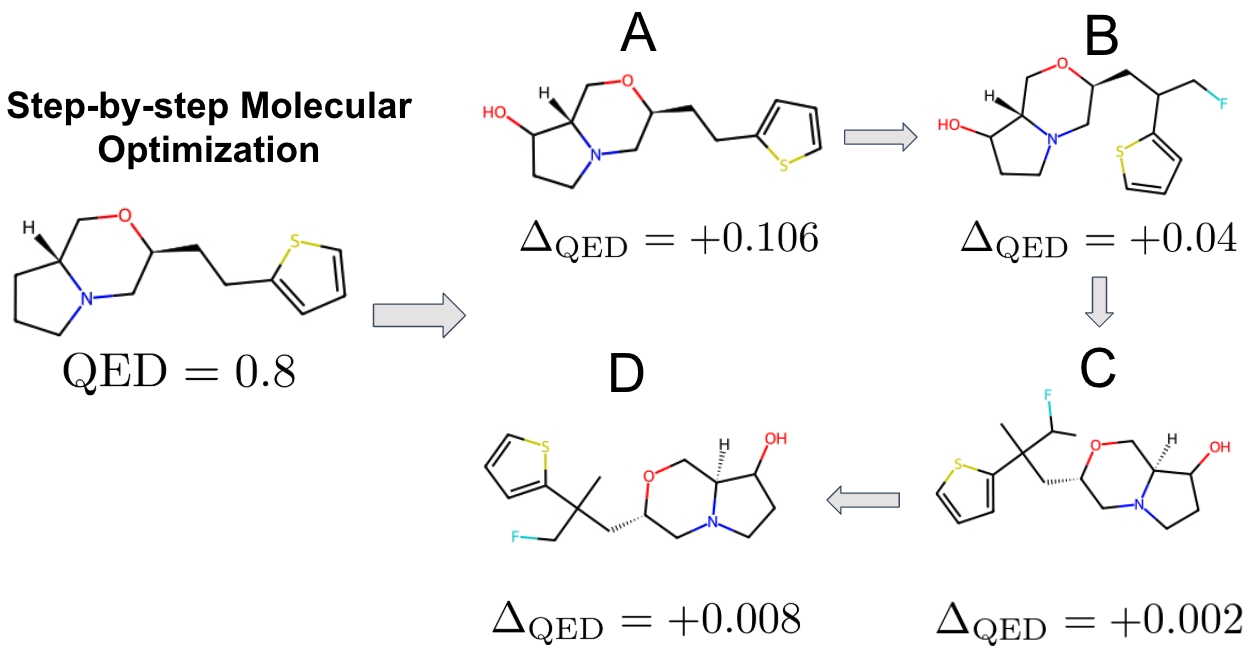 Given an existing molecule known to be effective against a certain disease (such as COVID-19), we develop methodologies to further enhance its effectiveness using our molecular editing framework, which combines generative models with optimization techniques, called Latent Space Bayesian Optimization. In the plot, QED refers to the molecule’s drug-likeness score—an estimate of its potential to be used as a drug. By performing small, targeted modifications at each step, we aim to maximize this value.
Given an existing molecule known to be effective against a certain disease (such as COVID-19), we develop methodologies to further enhance its effectiveness using our molecular editing framework, which combines generative models with optimization techniques, called Latent Space Bayesian Optimization. In the plot, QED refers to the molecule’s drug-likeness score—an estimate of its potential to be used as a drug. By performing small, targeted modifications at each step, we aim to maximize this value.
To address that, I turned to Bayesian optimization, which is well-suited for making the most out of limited samples. Specifically, I built the method around latent space Bayesian optimization. Molecules are discrete and high-dimensional structures, so we use autoencoders to map them into a lower-dimensional, continuous latent space. This makes optimization much more manageable.
Instead of generating entirely new molecules from scratch, the model proposes small, targeted modifications — like adding small fragments to existing molecules — and even learns where to best attach them. This approach allows us to generate candidates that are not only optimized for desired properties but are also realistic and easier for chemists to synthesize.
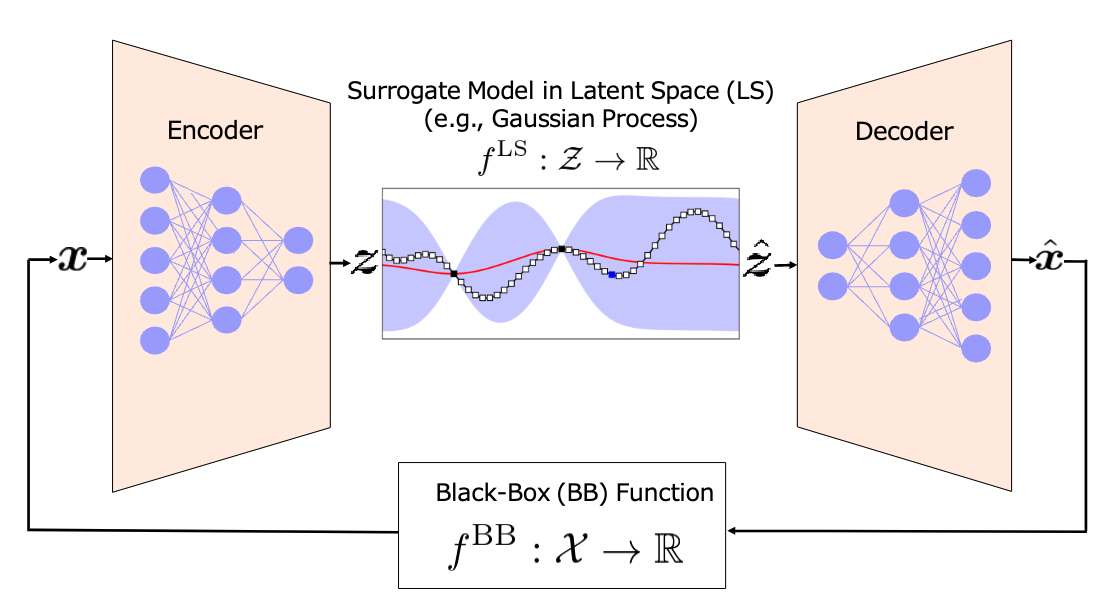 An illustration of Latent Space Bayesian Optimization (LSBO). LSBO is a technique for optimizing expensive black-box functions (such as the effectiveness of a molecule against a protein) by working in a simplified, compressed space created by a generative model like a Variational Autoencoder (VAE). First, an input example is encoded into a compact representation, called a latent vector, using the VAE’s encoder. A surrogate model (such as a Gaussian Process) is trained using these latent vectors and their corresponding function values. Bayesian optimization is then performed in this latent space to suggest a new promising latent vector. This is decoded back into a new input example, which is evaluated using the original black-box function. The process repeats, allowing efficient exploration and optimization in the latent space rather than directly in the complex input space.
An illustration of Latent Space Bayesian Optimization (LSBO). LSBO is a technique for optimizing expensive black-box functions (such as the effectiveness of a molecule against a protein) by working in a simplified, compressed space created by a generative model like a Variational Autoencoder (VAE). First, an input example is encoded into a compact representation, called a latent vector, using the VAE’s encoder. A surrogate model (such as a Gaussian Process) is trained using these latent vectors and their corresponding function values. Bayesian optimization is then performed in this latent space to suggest a new promising latent vector. This is decoded back into a new input example, which is evaluated using the original black-box function. The process repeats, allowing efficient exploration and optimization in the latent space rather than directly in the complex input space.
How does the project work in terms of collaboration with the chemists?
This project works in a kind of cycle. The chemists start by sending us some target information — basically what they’re aiming for — and then we generate a set of candidate molecules based on that. They pick a few to synthesize, and if one turns out to be promising, they might ask us to improve it further. In the next round, our algorithm takes that molecule as input and tries to optimize it even more.
Then we send them the updated candidates, and they move on to the next round of synthesis. This cycle keeps going until the chemists are happy with the result. Each cycle usually takes at least four months, so it’s a long process, but it’s collaborative and iterative, which I think is one of the strengths of the project.
I understand that you are due to finish your PhD quite soon. Have you got a position lined up for when you graduate?
Yeah, I’m actually in the middle of writing my thesis right now. I plan to submit it in about two months and defend it around August. After that, I’ll be starting a position at IBM Research in Tokyo. I interned there at the end of last year, and it was a great experience — I learned a lot, and that led to this opportunity to join them full-time after graduation.
Right now, I’m based in Nagoya, which is in central Japan. It’s just about an hour and a half to Tokyo by bullet train, so I’ll be moving there around August or September. The project I’ll be working on is closely related to my current research. I’ll be joining the materials discovery team, and we’ll be developing property prediction models for chemical compounds and materials — so it’s a nice continuation of what I’ve been doing during my PhD.
Moving on to talk about the AAAI conference. How was the Doctoral Consortium, and the conference experience in general?
It was actually my first time attending such a big conference — and also my first time in the U.S. — so the whole experience was really exciting. The Doctoral Consortium was super well-organized. In the mornings, we had mentoring sessions with professors and researchers from both academia and industry. They were really generous with their advice and shared a lot of useful insights from their own experiences.
It was also great to meet other PhD students. Most of us were at a similar stage — getting close to finishing — and it was honestly a relief to hear that everyone’s dealing with the same uncertainties about what comes next. Beyond research, we also talked a lot about the psychological side of doing a PhD. Personally, I think it’s more of a mental challenge than a purely academic one, so being able to openly talk about that was really valuable.
There were also some nice social events — shared lunches and a dinner — where we could connect more casually. During the poster session, I had some great conversations with people who were genuinely interested in my work, and it was fun trying to understand what others were working on too. The range of topics was super diverse, and I came away from it with a lot of new ideas. I really enjoyed the program and have already recommended it to everyone in my lab.
Aside from the Doctoral Consortium, I also had another paper accepted to AAAI — this one at the “AI to Accelerate Science and Engineering” workshop. Unfortunately, I could not join the workshop sessions due to a conflict with another conference.
As for the main conference, I got to attend some of the poster sessions and keynote talks. It was inspiring and also gave me some direction for the future. A lot of people are working on large language models and AI reasoning now, so I’m starting to think about how to bring those ideas into my own research, especially in the context of scientific discovery.
How did you find that AlphaFold changed the research landscape in your area?
AlphaFold has actually had a big impact on our work. In our molecule design projects, we’re focused on creating compounds that can interact well with target proteins — so having good binding affinity is key. We’re not doing wet lab experiments ourselves; everything happens in a simulated environment, and those simulations rely on predicted 3-dimensional protein structures. That’s where AlphaFold comes in — we use the structures it predicts as input for our optimization process.
I’m not working directly on AlphaFold — that part is handled by other researchers — but our results really depend on how accurate those predictions are. If AlphaFold gives us poor protein structures, our molecule designs won’t be very effective either.
Overall, AlphaFold has really pushed the field forward. It’s made protein structure prediction more accessible, helped our area grow, and also brought in more attention and funding, which has definitely helped.
Final question, do you have any interesting hobbies or interests outside of AI?
Yeah, I’m really into Formula One — I try to follow the races whenever I can, and I also do a bit of sim racing myself. I also really enjoy boxing. I used to box when I was younger, and I picked it up again during my first two years in Japan, but then I got injured, so I had to take a break. I’m hoping to make a comeback soon though! Earlier I mentioned how a PhD can be mentally challenging, and I feel like boxing is the same — it’s not just about physical strength. It’s also about how well you can stay calm, read your opponent, and manage your emotions in a really intense setting. It’s just you and the other person in a small space — very psychological. That’s one of the things I love about it.
About Onur

|
Onur Boyar is a PhD candidate at Nagoya University in Japan and a researcher at RIKEN. His research focuses on the design of novel molecules and crystals using generative models, latent space representations, and advanced optimization techniques such as Bayesian optimization. Prior to his doctoral studies, he worked as a data scientist at several companies, gaining hands-on experience in applied machine learning. He got his Bachelor’s and Master’s degrees from Boğaziçi University, Turkey. Onur is passionate about leveraging AI to accelerate scientific discovery, particularly in the fields of chemistry and materials science. |












![Life in Startup Pivot Hell with Ex-Microsoft Lonewolf Engineer Sam Crombie [Podcast #171]](https://cdn.hashnode.com/res/hashnode/image/upload/v1746753508177/0cd57f66-fdb0-4972-b285-1443a7db39fc.png?#)






_Andrey_Khokhlov_Alamy.jpg?width=1280&auto=webp&quality=80&disable=upscale#)